A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) | Semantic Scholar

Quantitative proteomics - SILAC Harini Chandra a The identification and quantitation of complex protein mixtures have been facilitated by mass spectrometric. - ppt download

Schematic representation of a stable isotope labelling with amino acids... | Download Scientific Diagram

Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) for Studying Dynamics of Protein Abundance and Posttranslational Modifications

An Enhanced In Vivo Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) Model for Quantification of Drug Metabolism Enzymes * - Molecular & Cellular Proteomics
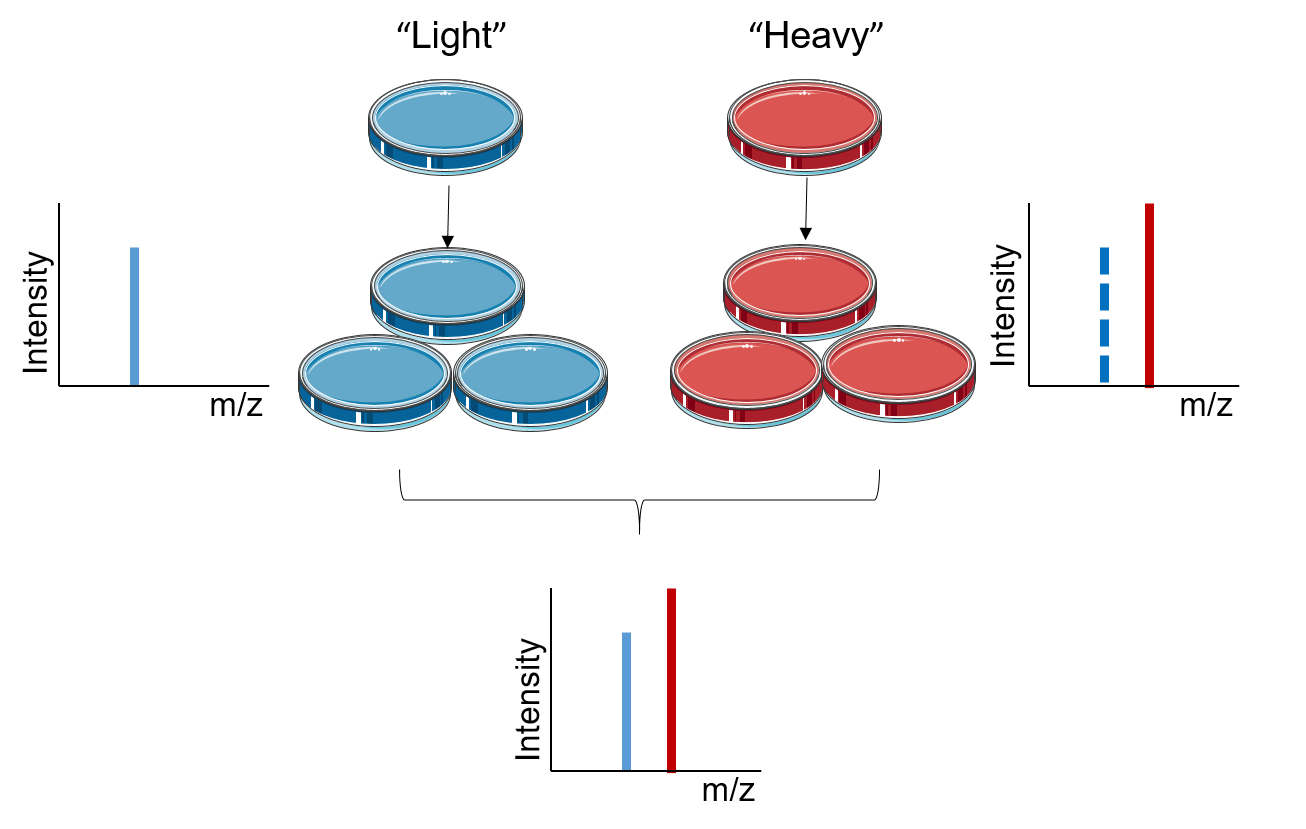
Stable Isotope Labeling Using Amino Acids in Cell Culture (SILAC): Principles, Workflow, and Applications – Creative Proteomics Blog
Chain Polypeptide Stock Illustrations – 222 Chain Polypeptide Stock Illustrations, Vectors & Clipart - Dreamstime

Comparison of Isotope-labeled Amino Acid Incorporation Rates (CILAIR) Provides a Quantitative Method to Study Tissue Secretomes - Molecular & Cellular Proteomics
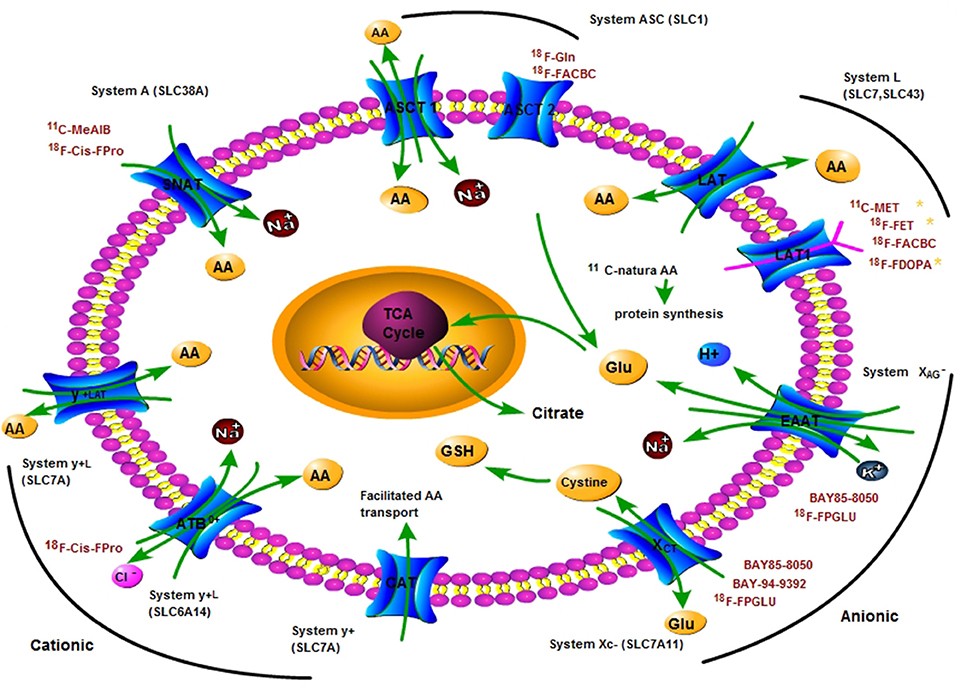
Frontiers | Carbon-11 and Fluorine-18 Labeled Amino Acid Tracers for Positron Emission Tomography Imaging of Tumors | Chemistry
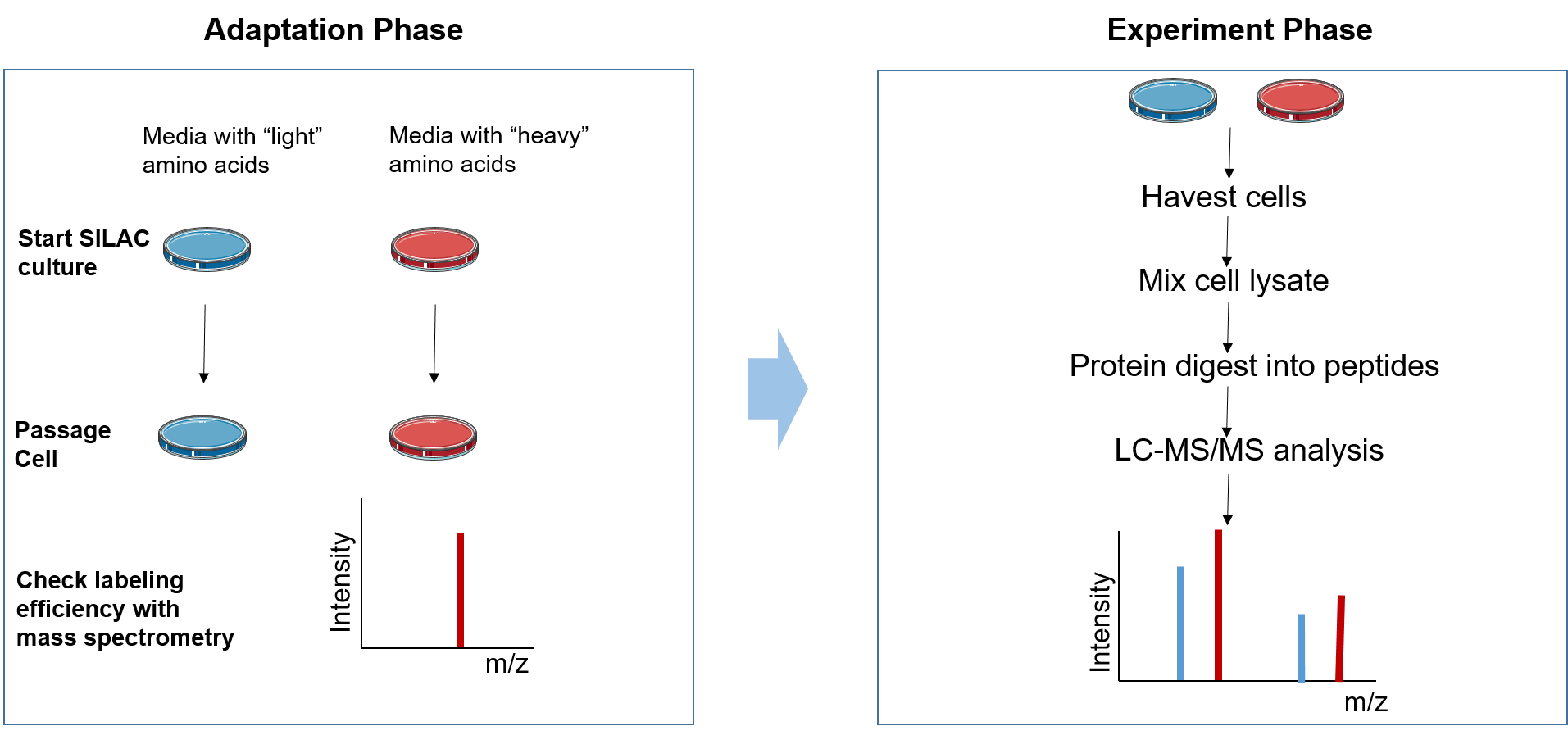
Stable Isotope Labeling Using Amino Acids in Cell Culture (SILAC): Principles, Workflow, and Applications – Creative Proteomics Blog

Intercellular Transfer of Proteins as Identified by Stable Isotope Labeling of Amino Acids in Cell Culture* - Journal of Biological Chemistry