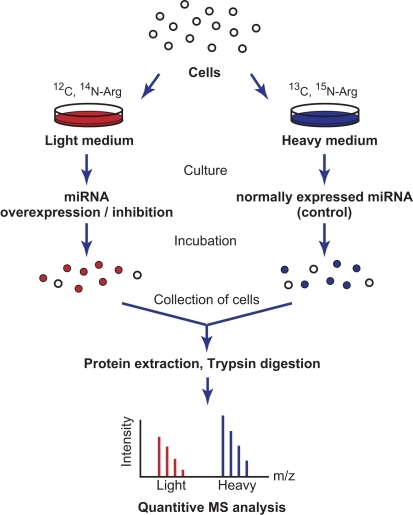
IJMS | Free Full-Text | SILAC-Based Quantification of TGFBR2-Regulated Protein Expression in Extracellular Vesicles of Microsatellite Unstable Colorectal Cancers | HTML

Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) for Studying Dynamics of Protein Abundance and Posttranslational Modifications

Intercellular Transfer of Proteins as Identified by Stable Isotope Labeling of Amino Acids in Cell Culture* - Journal of Biological Chemistry
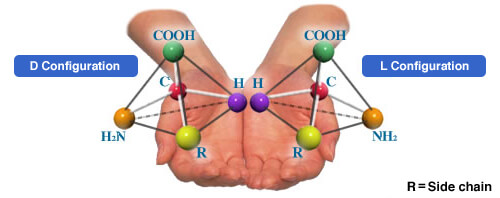
What are L-, D-, and DL-Amino Acids? | Enhancing Life with Amino Acids | About us | Ajinomoto Group Global Website - Eat Well, Live Well.

Figure 3 | Proteomics progresses in microbial physiology and clinical antimicrobial therapy | SpringerLink

Light or heavy isotope labeled amino acids (Lys, +6 Da; Arg, +10 Da)... | Download Scientific Diagram

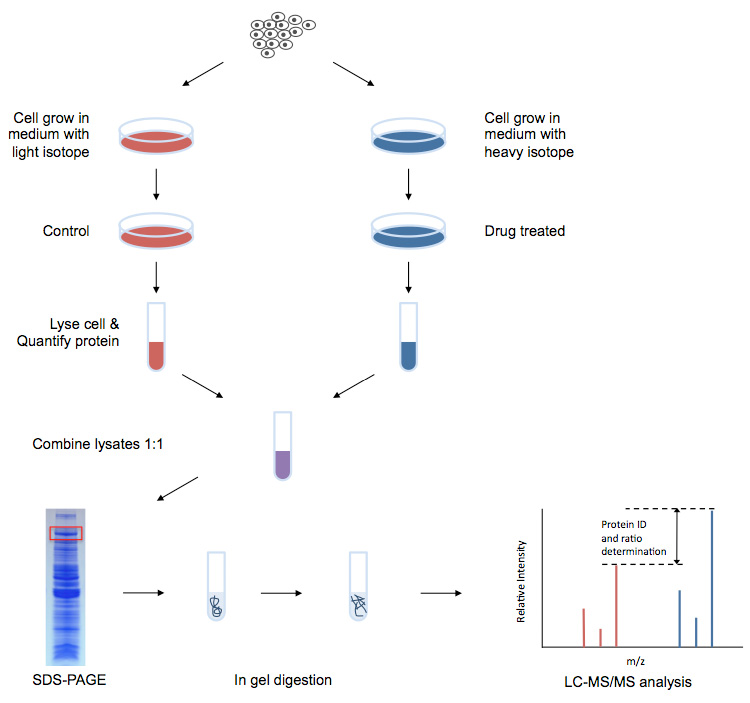
Characterization of the Tyrosine Kinase-Regulated Proteome in Breast Cancer by Combined use of RNA interference (RNAi) and Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) Quantitative Proteomics* - Molecular &

Cells can be metabolically labeled with a combination of Aha and/or... | Download Scientific Diagram

Amino acid based labeling techniques. (A) SILAC labeling. Proteins are... | Download Scientific Diagram

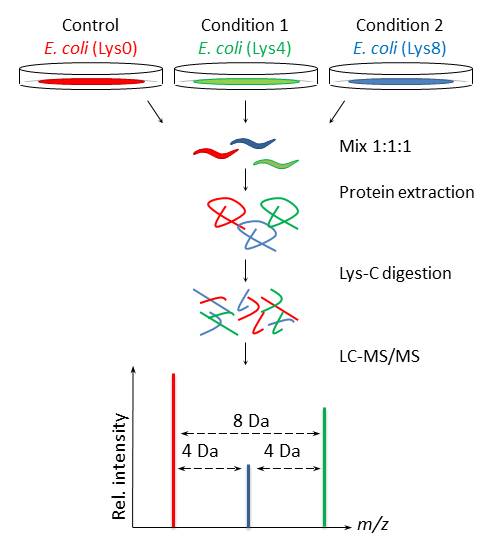
Comparison of Isotope-labeled Amino Acid Incorporation Rates (CILAIR) Provides a Quantitative Method to Study Tissue Secretomes - Molecular & Cellular Proteomics

Encoding quantitative information into whole proteomes with SILAC.Cells... | Download Scientific Diagram

Quantitative, Time-Resolved Proteomic Analysis by Combining Bioorthogonal Noncanonical Amino Acid Tagging and Pulsed Stable Isotope Labeling by Amino Acids in Cell Culture* - Molecular & Cellular Proteomics

Light or heavy isotope labeled amino acids (Lys, +6 Da; Arg, +10 Da)... | Download Scientific Diagram

Schematic representation of a stable isotope labelling with amino acids... | Download Scientific Diagram